Entity Extraction API
In this tutorial, we will employ the Entity Extraction API to convert unstructured clinical data, (e.g., History & Physical notes), into structured data elements.
The Entity Extraction API normalizes unstructured free text in real-time. The identified terms include rich metadata mappings to multiple coding systems (e.g., SNOMED-CT, ICD-10-CM, etc.).
The API is intended to be invoked with single records at a time, for example, the text from clinical notes for a single note.
Use Cases
The Entity Extraction API uses Named entity recognition (NER) to identify problem/diagnosis, procedure, medication, and laboratory terms. The API addresses issues associated with unstructured data, solving for a number of use cases:
- Decision support and analytics
- Improved revenue workflows
- Improved patient care
- Comprehensive clinical data with NER
- Coordination with multiple coding systems
- Quality and consistency
Let’s discuss: What problems can be addressed with the Entity Extraction API?
We estimate that having the capability to transform unstructured clinical data into structured data elements increases the ability to extract clinical meaning from the data by ~10%. In addition to History & Physical notes, unstructured data can come in the form of Discharge Summary Narratives, Progress Notes, and Procedure Notes.
Think Deeper: Get better acquainted with the use cases applicable to the Entity Extraction API. Go to the Topic Spotlight section to learn more about the use cases, as well as other details associated with the API.
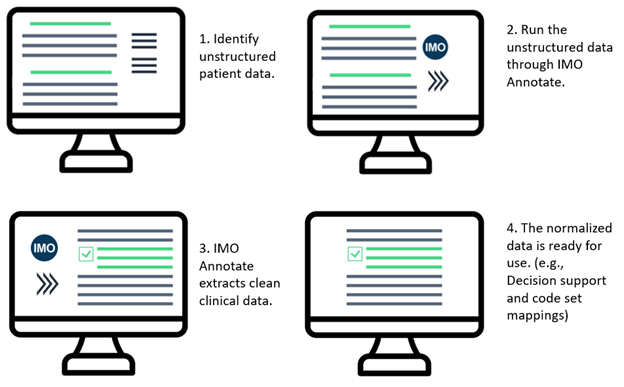
Setup
Start with a record of unstructured free text.
NLP Example Text:
Chief Complaint: “chest pain” HPI: -- is a 76 yo man with h/o HTN, DM, and sleep apnea who presented to the ED complaining of chest pain. He states that the pain began the day before and consisted of a sharp pain that lasted around 30 seconds, followed by a dull pain that would last around 2 minutes. The pain was reported as left chest pain. The onset of pain came while the patient was walking in his home. He did not sit and rest during the pain but continued to do household chores. Later on in the afternoon he went to the gym where he walked 1 mile on the treadmill, rode the bike for 5 minutes, and swam in the pool. He did not have any reoccurrences of chest pain while at the gym or later in the evening. The following morning (of his presentation to the ED) he noticed the pain as he was getting out of bed. Once again it was a dull pain, preceded by a short interval of a sharp pain. The patient did experience some tingling in his right arm after the pain ceased. He continued to have several episodes of the pain throughout the morning, so his daughter-in-law decided to take him to the ED around 12:30pm. The painful episodes did not increase in intensity or severity during this time. At the ED the patient was given nitroglycerin, which he claims helped alleviate the pain somewhat. -- has not experienced any shortness of breath, no nausea, or no diaphoresis during these episodes of pain. He has never had chest pain in the past. He was told “years ago” that he has a right bundle branch block and premature heart beats. Procedure History: he had an appendectomy in 2002.
API Usage
The API can be accessed at: https://api.imohealth.com/entityextraction/pipelines.
An IMO Precision Normalize agreement is necessary to receive authentication credentials.
Following entering an agreement, API keys are provided and can be used to generate tokens. For more information, contact IMO Customer Support.
Authentication
Authentication is performed using OAuth 2.0 Bearer tokens which requires an IMO-provided client ID and secret.
The following code snippets demonstrate retrieving auth tokens:
curl
curl -u "<API Key>:<API Secret>" \
--data "grant_type=client_credentials&audience=https://api.imohealth.com" \
"https://api.imohealth.com/oauth/token"python
import requests
data = {
'grant_type':'client_credentials',
'client_id':'<API_KEY>',
'client_secret':'<API_SECRET>',
'audience':'https://api.imohealth.com'
}
response = requests.post(
url = "https://api.imohealth.com/oauth/token",
json = data
)
response = response.json()
auth_token = response['access_token']
print(auth_token)
How to call the Entity Extraction API
The Entity Extraction API leverages natural language processing (NLP) models for the identification of clinically relevant terms within unstructured text.
Description: This API will extract and classify Problem, Procedure, Medication, and Lab terms from the unstructured clinical text provided. These terms will include mappings to external vocabularies, such as SNOMED and ICD.
This functionality can be reached via a POST request to the /entityextraction/pipelines/{pipeline_name}?version={pipeline_version} HTTP endpoint, as shown in the examples below:
curl
curl -X POST \
-H "Authorization: Bearer <AUTH_TOKEN>" \
-H "Content-Type: application/json" \
-data "{'text': 'John Smith has a headache and reports pain in his left leg.'}"\
"https://api.imohealth.com/entityextraction/pipelines/imo-clinical-comprehensive?version=3.0" python
import requests
auth_token='<AUTH_TOKEN>'
headers = {'Authorization': 'Bearer ' + auth_token}
data = {'text': 'HISTORY OF PRESENT ILLNESS\nJohn Smith has a headache and reports pain which is in his left leg.', 'preferences': {'type_filter': ['entities', 'relations']}}
response = requests.post(
url = https://api.imohealth.com/entityextraction/pipelines/imo-clinical-comprehensive?version=3.0
json = data,
headers = headers
}
print(response.json())Response
{
"filename": "",
"content": "HISTORY OF PRESENT ILLNESS\nJohn Smith has a headache and reports pain which is in his left leg.",
"entities": [
{
"id": "44_52_Entity_problem",
"text": "headache",
"begin": 44,
"end": 52,
"type": "standard",
"semantic": "problem",
"section": "history_present_illness",
"assertion": "present",
"explanation": "Entity found at position 44 to 52 of input text.",
"codemaps": { "imo": { "lexical_title": "Headache", "lexical_code": "45536", "default_lexical_title": "Headache", "default_lexical_code": "45536", "confidence": "1.0" }, ... "icd10cm": {"codes": [ { "code": "R51.9", "title": "Headache, unspecified", "map_type": "Preferred primary", ... } ] }, ...},
},
{
"id": "65_69_Entity_problem",
"text": "pain",
"begin": 65,
"end": 69,
"type": "standard",
"semantic": "problem",
"section": "history_present_illness",
"assertion": "present",
"explanation": "Entity found at position 65 to 69 of input text.",
"linked_entities": [
{
"id": "86_94_Entity_bodyloc",
"text": "left leg"
}
],
"codemaps": { "imo": { "lexical_title": "Pain of left lower extremity", "lexical_code": "48471000", "default_lexical_title": "Pain of left lower extremity", "default_lexical_code": "48471000", "confidence": "1.0" }, ... "icd10cm": { "codes": [ { "code": "M79.605", "title": "Pain in left leg", "map_type": "Preferred primary", ... } ] }, ... }
},
{
"id": "86_94_Entity_bodyloc",
"text": "left leg",
"begin": 86,
"end": 94,
"type": "standard",
"semantic": "bodyloc",
"section": "history_present_illness",
"assertion": "present",
"explanation": "Entity found at position 86 to 94 of input text."
}
],
"relations": [
{
"id": "65_69_Entity_problem_86_94_Entity_bodyloc_Relation_problem-bodyloc",
"begin": 65,
"end": 94,
"semantic": "problem-bodyloc",
"from_ent": "65_69_Entity_problem",
"from_ent_text": "pain",
"to_ent": "86_94_Entity_bodyloc",
"to_ent_text": "left leg"
}
],
"text_id": "fef87500-c5d1-42f6-aa05-a83ea7316814",
"pipeline": {
"name": "imo-clinical-comprehensive",
"version": "3.0"
},
"preferences": {
"thresholds": {
"problem": 0.85,
"procedure": 0.85,
"medication": 0.85,
"lab": 0.85
},
"type_filter": [
"entities",
"relations"
],
"domain_filter": [
"problem",
"procedure",
"medication",
"lab"
]
},
"metadata": {}
}(Note: The response above is truncated to display a brief selection of meaningful normalization. To view the full schema of the response, see the Reference Documentation tab for Entity Extraction.)
Parsing the NLP JSON response
Our JSON response organizes identified NLP components by their type. Component types include:
- Tokens
- Sentences
- Entities
- Relations
To include or exclude component types, use the type_filter Preference option.
For a more comprehensive schema, see the Reference Documentation.
Understanding entity relationships
Semantically relevant relationships between entities are represented in two ways. First, relationships between two individual entities are represented within a Relation collection. Each relation contains a from entity and a to entity, as well as a description of the type of relationship (semantic).
Example
"relations": [
{
"id": "38_42_Entity_problem_59_67_Entity_bodyloc_Relation_problem-bodyloc",
"begin": 38,
"end": 67,
"semantic": "problem-bodyloc",
"from_ent": "38_42_Entity_problem",
"from_ent_text": "pain",
"to_ent": "59_67_Entity_bodyloc",
"to_ent_text": "left leg"
}
] The second-place relationships can be found is within the linked_entities property of an Entity. This property captures the IDs and text of all entities related to this “base” entity. These relationships are also considered when providing codemaps for the entities.
Example
{
"id": "66_70_Entity_problem",
"text": "pain",
"begin": 66,
"end": 70,
"type": "standard",
"semantic": "problem",
"section": "history_present_illness",
"assertion": "present",
"explanation": "Entity found at position 66 to 70 of input text.",
"linked_entities": [
{
"id": "78_86_Entity_bodyloc",
"text": "left leg"
}
],
"codemaps": {...} // code mappings for this entity with its relations
}
Summary
You sent a call to the Entity Extraction API with a goal to extract computable, clinician-friendly IMO terms from clinical unstructured free text. The API used NER to extract and classify Problem, Procedure, Medication and Lab terms from the unstructured clinical text provided.
Our NLP Service takes a 3-step approach to getting you clinically relevant codes for your unstructured text.
- Identify clinical entities, which are shown in the
entitiesarray in the response. - Group clinically related entities together to form more comprehensive, post-coordinated clinical concepts. See the
linked_entitiesattribute of an Entity, which contains the ids of other related Entities. - Resolve the grouped entities to standard code sets (e.g., ICD, SNOMED, IMO). See
codemapsproperty of an Entity.
Topic Spotlight
- Use Cases – Think Deeper
- NLP Features
- Named Entity Recognition
- Roadmap
1. Use Cases – Think Deeper
The Entity Extraction API solves issues with unstructured data:
- Decision support and analytics: The API can unlock valuable unstructured data in databases and documents by extracting terms and normalizing essential concepts.
- Improved revenue workflows: The API provides a path to add specificity to unspecified data, giving health care facilities opportunities to optimize facility revenue. Using NLP, improvements are seen in coding and compliance use cases, along with fewer insurance denials due to incomplete coding.
- Improved patient care: The API can help clinicians to deliver better quality care with enriched clinical documentation, setting the stage for data-driven outcomes.
- Comprehensive clinical data: The API uses NER to tokenize unstructured clinical text into problem/diagnosis, procedure, medication, lab, SDOH and diagnosis history data. Access to a complete set of clinical data provides health care facilities with more comprehensive information about their patients, outcomes, and costs.
- Coordination with multiple coding systems: Thanks to its integration with Precision Normalize, the Entity Extraction API provides relevant standard codes for problems, procedures, medications, and labs identified within unstructured clinical documentation.
- Identifying problem terms within unstructured clinical documentation will provide users with access to IMO, ICD-10-CM, HCC (if available), ICD-9-CM, and SNOMED codes.
- Identifying procedure terms within unstructured clinical documentation will provide users with access to IMO, ICD-10-PCS, LOINC, CPT, hcpcs, and SNOMED codes.
- Identifying medication terms within unstructured clinical documentation will provide users with access to IMO, RxNorm, NDC, and CVX codes.
- Identifying lab terms within unstructured clinical documentation will provide users with access to IMO, LOINC, CPT, HCPCS, ICD-10-PCS, and SNOMED-CT codes.
- Quality and consistency: Providing normalization on terms found in unstructured text allows a user to validate structured data to report differences.
2. Named entity recognition
Named entity recognition (NER) is a form of natural language processing (NLP). The goal of NER is to extract and classify terms mentioned in unstructured text into clinical domains e.g., Problem, Procedure, Medication, and Lab domains.
3. Available Features
The Entity Extraction API identifies clinically relevant Problem, Procedure, Medication and Lab entities within unstructured clinical documentation. The API is also designed to go deeper with these capabilities:
- Identifies assertion status of clinical entities based on context providing the capability to identify negated clinical entities for a more accurate picture of the patient’s situation.
- Identifies the section that the entity was extracted from, which allows the user to customize their inclusion and exclusion criteria for location of terms.
- Identifies the laterality and anatomical site of clinical entities for improved specificity of extracted data, as required by payers and regulatory agencies.
- Identifies relationships between terms to normalize to a more specified pre-coordinated clinical terminology code.
- Identify temporal objects: Recognize time-oriented information (e.g., symptom duration), as noted in a set of medical data.
Notices
©️ 2025 Intelligent Medical Objects, Inc. All Rights Reserved.
CPT®️ copyright 2024 American Medical Association. All rights reserved.
SNOMED®️ and SNOMED CT®️ are registered trademarks of IHTSDO.
LOINC®️ is a registered United States trademark of Regenstrief Institute, Inc.
RxNorm is publicly available data courtesy of the U.S. National Library of Medicine (NLM), National Institutes of Health, Department of Health and Human Services.